Product Sheet CG1215
Description
BACKGROUND Nucleosome is the basic structure unit of chromatin. Each nucleosome is composed of an octamer of core histones (two each of H2A, H2B, H3 and H4), around which two super-helical turns (80 base pairs) of DNA are wrapped. The core histones within the nucleosome share a common structural motif, the histone fold, and non-structured N-termini. Epigenetic modifications, such as acetylation, phosphorylation, methylation, ubiquitination, and ADP ribosylation, are of the highly conserved N-terminal tail of core histones influence the genetic potential of DNA. Distinct patterns of histone modification are observed at specific chromosomal regions and affect various reactions on chromosomes (transcription, replication, repair, and recombination). Such histone modifications may modulate the interaction of histone and DNA directly at one hand; and some of these ‘epigenetic’ marks recruit proteins that regulate chromatin structure on the other hand. For example, Heterochromatin Protein 1 (HP1) binds to histone H3 when its lysine 9 residue has been tri-methylated by the methyltransferase Suv39h. Given the number of chromosomes per cell, the number of nucleosomes per chromosome, the number of different histone tails, and the number of different modifications, the potential complexity of regulation is immense, the histone code hypothesis has been proposed , according to which each combination of post-translational modifications on a histone tail has a specific function.1
It was shown that histone H3 lysine 56 (H3K56) is a frequent site of acetylation in fungal species. Bulk K56 acetylation peaks during S-phase, where it plays a role in the DNA damage response. H3K56 is acetylated by Rtt109, a distant homolog of the p300/CBP histone acetyltransferase, and which acts preferentially on free, but not nucleosomal, histones. In vitro, acetylation by Rtt109 is extremely inefficient in the absence of either of two histone chaperone cofactors, Asf1 and Vps75. Oddly, while Asf1 is required for detectable K56 acetylation in vivo, the major binding partner for Rtt109 in the cell is Vps75, whose deletion has little effect on bulk K56 acetylation levels.2 H3K56 acetylation opens chromatin and enables histone gene transcription, DNA replication, and DNA repair and prevents epigenetic silencing.3 It is a mark of newly replicated chromatin, but it has also been linked to replication-independent histone replacement.
REFERENCES
1. Joost, H.G. & Thorens, B.: Mol Membr Biol. 18:247-56, 2001
2. Huang, S. & Czech, M.: Cell Metabol. 5:237-52, 2007
3. Chang, L. et al: Mol. Med. 10:65-71, 2004
2. Huang, S. & Czech, M.: Cell Metabol. 5:237-52, 2007
3. Chang, L. et al: Mol. Med. 10:65-71, 2004
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
(Click to Enlarge)
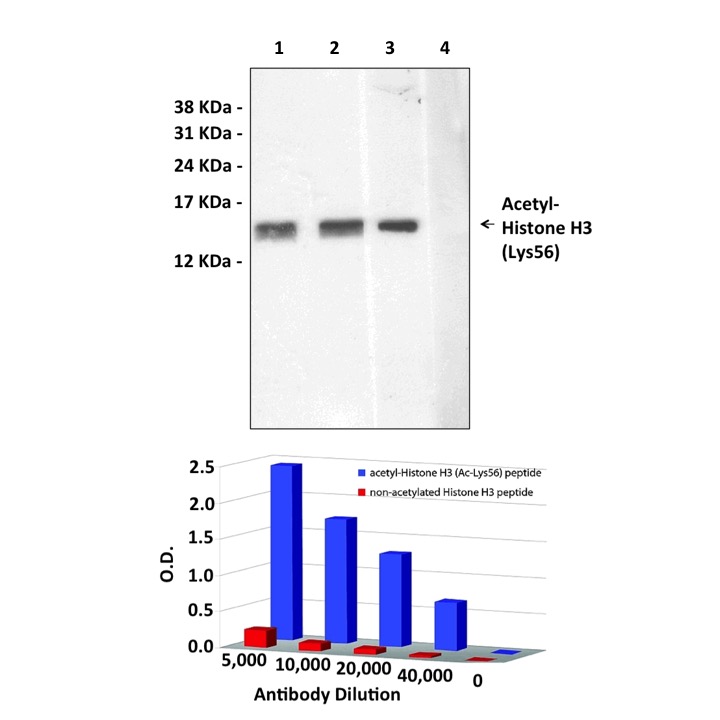
Top: Cell extracts were separated on SDS-PAGE, blotted with Anti-Acetyl-Histone H3 (Ac-Lys56) (antibody dilution 1:1,000), and developed using Goat Anti-Rabbit IgG-Peroxidase and a chemiluminescent substrate. Lanes: 1. HeLa 2. NIH3T3 3. NIH3T3 + non-acetylated Histone H3 peptide 4. NIH3T3 + immunizing acetylated Histone H3 (Ac-Lys56) peptide. Bottom: ELISA plates were coated with peptides at 0.5μg/mL, incubated with different dilutions of Anti-Acetyl-Histone H3 (Ac-Lys56), and developed using Goat Anti-Rabbit IgG-Alkaline Phosphatase, and a colorimetric substrate.
Details
Cat.No.: | CG1215 |
Antigen: | Synthetic peptide containing acetylated Lys56 of human histone H3, conjugated to KLH. The corresponding sequence is identical in many species including rat and mouse histone H3. |
Isotype: | Rabbit IgG |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
Applications & Suggested starting dilutions:* | WB 1:1000-1:2000 IP n/d IHC n/d ICC n/d FACS n/d |
Predicted Molecular Weight of protein: | 15 kDa |
Specificity/Sensitivity: | Detects endogenous Histone-H3 proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Rabbit Histone H3, Acetyl-Lys56 Antibody: Rabbit Histone H3, Acetyl-Lys56 Antibody | Size: 100 ul | CAT.#: CG1215 | Price: $384.00 |
