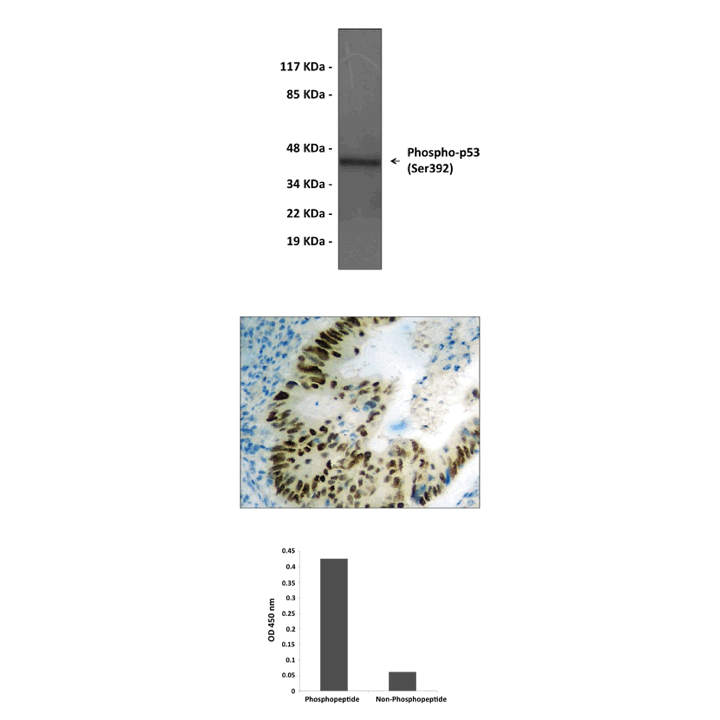
Product Sheet CG1388
Description
BACKGROUND The tumor suppressor p53 plays a critical role in preventing human cancer formation. In response to a variety of stress signals, often associated with the progression of neoplastic diseases, p53 becomes activated and induces cell cycle arrest and/or programmed cell death (apoptosis).1 By eliminating damaged and potentially dangerous cells that might otherwise become cancerous, p53 suppresses tumor formation. p53 has been called the "gatekeeper of the genome."
The p53 tumor suppressor protein is a tetrameric phosphoprotein and trasciptional factor.The 393-residue polypeptide contains four functional domains:N-terminal transcriptional activation domain (This is also the site where Mdm2 binds. Mdm2 binding is prevented by the phosphorylation of serine residues at positions 15 and 20); central DNA binding domain (most mutations occurred in this domain); the oligomerization domain ( contains nuclear localization signals and is involved in p53 tetramerization); and the basic C-terminal domain (residues 364-393) is a negative regulatory domain that can inhibit sequence-specific DNA binding by the core domain. Covalent modifications adding acetyl groups to lysine residues within this domain play a role in activating latent p53.1
In unstressed cells, p53 is latent and is maintained at low levels by targeted degradation mediated by its negative regulator, Mdm2. Mdm2 counteracts p53 tumor suppressor activity by physically binding to p53 and suppressing its transcriptional activity. Mdm2 also functions as the p53 ubiquitin ligase and triggers its degradation.2 Phosphorylations contribute to both the stabilization and activation of p53. For example, DNA-damaging agents activate phosphorylation at serine (Ser) 15, likely by a family of protein kinases including ATM and ATR, and Ser20 by the Chk2 kinase. These phosphorylation events are believed to contribute to p53 stabilization by preventing the binding of Mdm2 and rendering p53 more resistant to Mdm2. In addition, phosphorylation was also shown to modulate the transcriptional activity of p53. For example, phosphorylation at Ser15 stimulates p53 interaction with its transcriptional co-activators p300 and CBP, and a mutation that eliminates this phosphorylation leads to p53 transcriptional defects.3 Inappropriate expression viral or cellular oncogenes, such as ras or myc, leads to p53 activation through a p14ARF-dependent pathway. p14ARF inhibits the p53 ubiquitin ligase activity of Mdm2 and reduces degradation of p53. Another potential mechanism that may play a critical role in p53 activation is acetylation. Multiple lysine (K) residues in p53 are acetylated by p300 and its family member CBP or by P/CAF. Acetylation stimulates p53 DNA binding activity.4
The p53-dependent transcription of target genes responds to a diverse range of cellular signals that affect cell proliferation and DNA integrity checkpoints. Moreover, there are several potential mediators of p53-induced apoptosis. The Bax protein is an apoptosis-inducing member of the Bcl-2 protein family. Transcription of the BAX gene is directly activated by p53-binding sites in the regulatory region of the gene. Bax is located in mitochondria. When overexpressed, Bax induces apoptosis.5
The p53 tumor suppressor protein is a tetrameric phosphoprotein and trasciptional factor.The 393-residue polypeptide contains four functional domains:N-terminal transcriptional activation domain (This is also the site where Mdm2 binds. Mdm2 binding is prevented by the phosphorylation of serine residues at positions 15 and 20); central DNA binding domain (most mutations occurred in this domain); the oligomerization domain ( contains nuclear localization signals and is involved in p53 tetramerization); and the basic C-terminal domain (residues 364-393) is a negative regulatory domain that can inhibit sequence-specific DNA binding by the core domain. Covalent modifications adding acetyl groups to lysine residues within this domain play a role in activating latent p53.1
In unstressed cells, p53 is latent and is maintained at low levels by targeted degradation mediated by its negative regulator, Mdm2. Mdm2 counteracts p53 tumor suppressor activity by physically binding to p53 and suppressing its transcriptional activity. Mdm2 also functions as the p53 ubiquitin ligase and triggers its degradation.2 Phosphorylations contribute to both the stabilization and activation of p53. For example, DNA-damaging agents activate phosphorylation at serine (Ser) 15, likely by a family of protein kinases including ATM and ATR, and Ser20 by the Chk2 kinase. These phosphorylation events are believed to contribute to p53 stabilization by preventing the binding of Mdm2 and rendering p53 more resistant to Mdm2. In addition, phosphorylation was also shown to modulate the transcriptional activity of p53. For example, phosphorylation at Ser15 stimulates p53 interaction with its transcriptional co-activators p300 and CBP, and a mutation that eliminates this phosphorylation leads to p53 transcriptional defects.3 Inappropriate expression viral or cellular oncogenes, such as ras or myc, leads to p53 activation through a p14ARF-dependent pathway. p14ARF inhibits the p53 ubiquitin ligase activity of Mdm2 and reduces degradation of p53. Another potential mechanism that may play a critical role in p53 activation is acetylation. Multiple lysine (K) residues in p53 are acetylated by p300 and its family member CBP or by P/CAF. Acetylation stimulates p53 DNA binding activity.4
The p53-dependent transcription of target genes responds to a diverse range of cellular signals that affect cell proliferation and DNA integrity checkpoints. Moreover, there are several potential mediators of p53-induced apoptosis. The Bax protein is an apoptosis-inducing member of the Bcl-2 protein family. Transcription of the BAX gene is directly activated by p53-binding sites in the regulatory region of the gene. Bax is located in mitochondria. When overexpressed, Bax induces apoptosis.5
REFERENCES
1. Oren, M. : Biochem. Biophy. Acta 823:67-78, 1985
2. Waning, D.L. et al: Pharmaceuticals (Basel) 3:1576-93, 2010
3. Tanaka, T.: Seikagaku 82:200-9, 2010
4. Spange, S. et al: Int. J. Biochem. Cell Biol. 41:185-98, 2009
5. Farnebo, M. et al: Biocehm. Biophy. Res. Commun. 396:85-9, 2010
2. Waning, D.L. et al: Pharmaceuticals (Basel) 3:1576-93, 2010
3. Tanaka, T.: Seikagaku 82:200-9, 2010
4. Spange, S. et al: Int. J. Biochem. Cell Biol. 41:185-98, 2009
5. Farnebo, M. et al: Biocehm. Biophy. Res. Commun. 396:85-9, 2010
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Details
Cat.No.: | CG1388 |
Antigen: | Range AA363 to 393 |
Isotype: | Rabbit IgG |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
Applications & Suggested starting dilutions:* | WB 1:500-1:1000 IP n/d IHC 1:50-1:100 ICC n/d FACS n/d ELISA 1:1000 |
Predicted Molecular Weight of protein: | 43 kDa |
Specificity/Sensitivity: | Detects endogenous p53 proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Rabbit p53, Phospho-Ser392 Antibody: Rabbit p53, Phospho-Ser392 Antibody | Size: 100 ul | CAT.#: CG1388 | Price: $384.00 |