Product Sheet CG1626
Description
BACKGROUND UBF belongs to the sequence-nonspecific class of high mobility group (HMG) proteins and appears to function exclusively in Pol I transcription. UBF consists of two polypeptides (UBF1 and 2), which form hetero- and homodimers and arise from alternative splicing of a single transcript. Although UBF1 supports robust rRNA gene transcription, UBF2 is fivefold less active. In one model, UBF acts through its multiple HMG boxes to induce looping of DNA. This creates the enhancesome, a nucleosome-like structure thought to be responsible for the ability of UBF to modulate rRNA gene transcription. Early studies suggested that UBF1 acts predominantly at the promoter in the recruitment of SL1 (selectivity factor 1) and Pol I and in the formation of the preinitiation complex. More recently, additional roles have been ascribed to UBF1, including regulation of promoter escape and transcription elongation. Importantly, the association of UBF1 with rRNA genes in vivo is not restricted to the promoter but extends across the entire transcribed portion and to a lesser extent to the intergenic spacer (IGS). Indeed, consistent with its ability to modify DNA conformation, the inclusion of rRNA gene sequences with high affinity for UBF into ectopic sites on human chromosomes results in the formation of NOR-like structures indicative of “open” chromatin. In addition, targeting UBF1 to regions of heterochromatin is sufficient to induce large-scale chromatin decondensation. Furthermore, it was demonstrated that UBF regulates the open chromatin structure found in active rRNA genes by preventing linker histone H1–induced assembly of transcriptionally inactive chromatin. Long-term rRNA gene silencing in response to UBF depletion is stably propagated through the cell cycle and through many generations and is not associated with heterochromatic marks related to nucleolar remodeling complex (NoRC)–dependent remodeling, including DNA methylation. Restoring UBF levels rescues the number of active genes. Thus, in contrast to epigenetically silenced rRNA genes, which are methylated, silencing of rRNA genes through UBF depletion is reversible. Moreover, the pool of active ribosomal genes is not static but decreases during differentiation and that this decrease correlates with diminished UBF levels in the absence of changes in ribosomal DNA (rDNA) methylation.1 Thus, it is suggested that UBF1 binding throughout the rRNA gene repeat might contribute to the formation of the active chromatin state of rRNA genes. UPF plays important role in in chromatin remodeling.
Several studies have suggested roles for casein kinase II (CKII), cyclin-dependent kinase 2/4 (CDK2/4), mTOR/p70S6K and MAP kinase (ERK) in regulating UBF activity2,3 Thus, various signaling pathways induces an immediate upregulation of ribosomal transcription via UBF modification. It also demonstrates the existence of a direct link between growth factor signaling and ribosome biogenesis in mammalian cells. As cells enter mitosis, ribosomal gene transcription is shut down. It was shown both SL1 and UBF are inactivated by phosphorylation at mitosis and reactivated by dephosphorylation at the exit from mitosis and during G1 progression, respectively.4 Differentiation, serum deprivation, and glucocorticoids have been found to induce a regulation of PolI activity, probably via the polymerase-associated UBF or other factors. These regulations occur over extended time periods, reinforcing the established view that ribosomal transcription responds indirectly to changes in cellular metabolism.5
Several studies have suggested roles for casein kinase II (CKII), cyclin-dependent kinase 2/4 (CDK2/4), mTOR/p70S6K and MAP kinase (ERK) in regulating UBF activity2,3 Thus, various signaling pathways induces an immediate upregulation of ribosomal transcription via UBF modification. It also demonstrates the existence of a direct link between growth factor signaling and ribosome biogenesis in mammalian cells. As cells enter mitosis, ribosomal gene transcription is shut down. It was shown both SL1 and UBF are inactivated by phosphorylation at mitosis and reactivated by dephosphorylation at the exit from mitosis and during G1 progression, respectively.4 Differentiation, serum deprivation, and glucocorticoids have been found to induce a regulation of PolI activity, probably via the polymerase-associated UBF or other factors. These regulations occur over extended time periods, reinforcing the established view that ribosomal transcription responds indirectly to changes in cellular metabolism.5
REFERENCES
1. Sanij, E. et al: J. Cell Biol. 183:1259-74, 2008
2. Stefanovsky, V.Y. et al: Mol. Cell 8:1063-73, 2001
3. Hannan, K.M. et al: Mol. Cell. Biol. 23:8862-77, 2003
4. Klein, J. & Grumm, I.: Proc. Natl. Acad. Sci. USA 96:6096-101, 1999
5. Stefanovsky, V.Y. et al: Mol. Cell 21:629-39, 2006
2. Stefanovsky, V.Y. et al: Mol. Cell 8:1063-73, 2001
3. Hannan, K.M. et al: Mol. Cell. Biol. 23:8862-77, 2003
4. Klein, J. & Grumm, I.: Proc. Natl. Acad. Sci. USA 96:6096-101, 1999
5. Stefanovsky, V.Y. et al: Mol. Cell 21:629-39, 2006
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
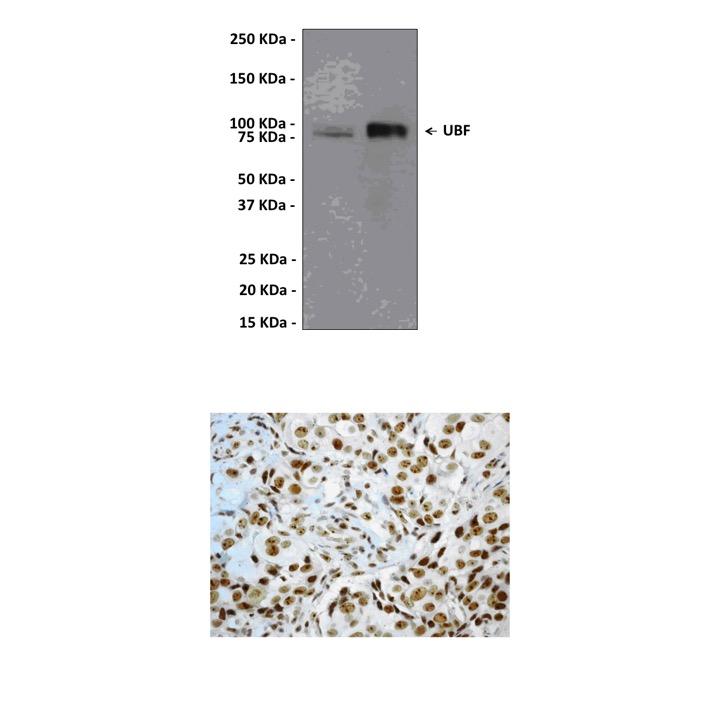
(Click to Enlarge) Top: Western Blot detection of UBF proteins in various cell lysates using UBF Antibody.HeLa (left lane) and A431 (right lane) cell lysates were probed with Anti-UBF, clone EP2741Y (1:1,000-10,000 dilution). Proteins were visualized using a Goat Anti-Rabbit secondary antibody conjugated to HRP and detected using a chemiluminescence detection system. Bottom: Immunohistochemical analysis of paraffin-embedded human breast carcinoma using Anti-UBF, clone EP2741Y.
Details
Cat.No.: | CG1626 |
Antigen: | Synthetic peptide corresponding to human UBF near the N-terminus. |
Isotype: | Rabbit IgG |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
Applications & Suggested starting dilutions:* | WB 1:1000-1:10000 IP n/d IHC n/d ICC 1:100-1:250 FACS n/d |
Predicted Molecular Weight of protein: | 94 kDa |
Specificity/Sensitivity: | This antibody recognizes UBF near the N-terminus. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Rabbit Upstream Binding Factor Antibody: Rabbit Upstream Binding Factor Antibody | Size: 100 ul | CAT.#: CG1626 | Price: $333.00 |
