Product Sheet CP10237
Description
BACKGROUND Tat-Interactive Protein 60 (TIP60) is a member of the MYST family proteins, which are related by an atypical Histone Acetyltransferase (HAT) domain. It has been shown that TIP60 has divergent functions and roles in many processes, such as cellular signaling, DNA damage repair, cell cycle and checkpoint control and apoptosis. It is a multifunctional enzyme involved in multiple nuclear transactions. The TIP60 regulates the DNA damage response following genotoxic stress by acetylating histone and remodeling chromatin. Indeed, in response to DNA double strand breaks, TIP60 is recruited to DNA lesions where it participates both in the initial as well as the final stages of repair. The ATM and DNA-PKcs activation is dependent on its acetylation by TIP60 in response to DNA double break repair, indicating that TIP60 plays crucial roles in DNA damage response. Indeed, mounting evidence implies that TIP60 functions at multiple levels in DNA damage repair, and its histone acetyltransferase activity acts as an upstream component in DNA damage checkpoint activation. Additionally, acetylation of lysine 120 of p53 by TIP60 may help distinguish the cell-cycle arrest and apoptotic functions of p53.2
Besides histone acetylase activity on chromatin remodeling, recent data indicate that TIP60 has more divergent functions in many processes. In transcriptional regulation, TIP60 is a tightly regulated transcriptional coregulator, acting in a large multiprotein complex for a range of transcription factors including androgen receptor, Myc, STAT3, NF-kappaB, E2F1 and p53. This usually involves recruitment of TIP60 acetyltransferase activities to chromatin. For instance, androgen receptor (AR) is acetylated by TIP60, which enhances its trans-activation in a ligand-dependent manner. Interaction of TIP60 with BLC-3 regulates the transcriptional activity of NF-kappaB; The ability of c-MYC to activate transcription relies in part on the recruitment of cofactor complexes containing TIP60, which directly acetylates c-MYC.3 Futhermore, regulation of TIP60 by post-translational modification has been documented. Similar to p53, TIP60 is subjected to proteasome-dependent proteolysis by interacting physically with Mdm2, a specific E3 ligase in ubiquitination cascade and accumulates following UV irradiation, indicating that ubiquitination of TIP60 could be part of the mechanism in tumorigenesis induced by Mdm2 and p53 following cellular stress. Moreover, it has been revealed that TIP60 can be phosphorylated at Ser86 and Ser90, whereas TIP60 histone acetyltransferase activity is controlled by its phosphorylation. Specifically, the phosphorylation of Ser90 is modulated by cyclin B/Cdc2; accordingly, phosphorylated TIP60 is accumulated after drug-induced arrest of cells in G2/M or at the G2/M transition, which strongly suggests a cell cycle-dependent control of TIP60 activity. As a histone acetyltransferase, TIP60 can also be acetylated by p300/CREB-binding protein in the zinc finger domain at Lys268 and Lys282. Furthermore, it was shown that TIP60 is autoacetylated in response to UV damage, which is critically important for TIP60 activation. Mechanistically TIP60 autoacetylation leads to the dissociation of TIP60 oligomer and enhances its interaction with substrates. Moreover, SIRT1 specifically deacetylates TIP60 and negatively regulates TIP60 activity in vivo.4 Additionally,it was shown that TIP60 is a novel substrate of sumoylation. The sumoylation of TIP60 augments its acetyltransferase activity in vitro and in vivo.5 Moreover ATF2 in cooperation with Cul3 ubiquitin ligase promotes degradation of TIP60.6 Finally, monoallelic loss of TIP60 was found in human carcinomas.
Besides histone acetylase activity on chromatin remodeling, recent data indicate that TIP60 has more divergent functions in many processes. In transcriptional regulation, TIP60 is a tightly regulated transcriptional coregulator, acting in a large multiprotein complex for a range of transcription factors including androgen receptor, Myc, STAT3, NF-kappaB, E2F1 and p53. This usually involves recruitment of TIP60 acetyltransferase activities to chromatin. For instance, androgen receptor (AR) is acetylated by TIP60, which enhances its trans-activation in a ligand-dependent manner. Interaction of TIP60 with BLC-3 regulates the transcriptional activity of NF-kappaB; The ability of c-MYC to activate transcription relies in part on the recruitment of cofactor complexes containing TIP60, which directly acetylates c-MYC.3 Futhermore, regulation of TIP60 by post-translational modification has been documented. Similar to p53, TIP60 is subjected to proteasome-dependent proteolysis by interacting physically with Mdm2, a specific E3 ligase in ubiquitination cascade and accumulates following UV irradiation, indicating that ubiquitination of TIP60 could be part of the mechanism in tumorigenesis induced by Mdm2 and p53 following cellular stress. Moreover, it has been revealed that TIP60 can be phosphorylated at Ser86 and Ser90, whereas TIP60 histone acetyltransferase activity is controlled by its phosphorylation. Specifically, the phosphorylation of Ser90 is modulated by cyclin B/Cdc2; accordingly, phosphorylated TIP60 is accumulated after drug-induced arrest of cells in G2/M or at the G2/M transition, which strongly suggests a cell cycle-dependent control of TIP60 activity. As a histone acetyltransferase, TIP60 can also be acetylated by p300/CREB-binding protein in the zinc finger domain at Lys268 and Lys282. Furthermore, it was shown that TIP60 is autoacetylated in response to UV damage, which is critically important for TIP60 activation. Mechanistically TIP60 autoacetylation leads to the dissociation of TIP60 oligomer and enhances its interaction with substrates. Moreover, SIRT1 specifically deacetylates TIP60 and negatively regulates TIP60 activity in vivo.4 Additionally,it was shown that TIP60 is a novel substrate of sumoylation. The sumoylation of TIP60 augments its acetyltransferase activity in vitro and in vivo.5 Moreover ATF2 in cooperation with Cul3 ubiquitin ligase promotes degradation of TIP60.6 Finally, monoallelic loss of TIP60 was found in human carcinomas.
REFERENCES
1. Squatrito, M.: Trends Cell Biol. 16:433-42, 2006
2. Sykes, S.M. Et al: Mol. Cell 24:841-51, 2006
3. Sapountzi, V. et al: Int. J.Biochem. Cell Biol. 38:1496:1509,2006
4. Wang, J. & Chen, J.: J. Biol. Chem. 285:11458-65, 2010
5. Cheng, Z. et al: Oncogene 27:931-47, 2008
6. Bhoumik, A. et al: J. Biol. Chem. 283:17605-14, 2008
2. Sykes, S.M. Et al: Mol. Cell 24:841-51, 2006
3. Sapountzi, V. et al: Int. J.Biochem. Cell Biol. 38:1496:1509,2006
4. Wang, J. & Chen, J.: J. Biol. Chem. 285:11458-65, 2010
5. Cheng, Z. et al: Oncogene 27:931-47, 2008
6. Bhoumik, A. et al: J. Biol. Chem. 283:17605-14, 2008
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Details
Cat.No.: | CP10237 |
Antigen: | Purified recombinant human TIP60 fragments expressed in E. coli. |
Isotype: | Mouse IgG2b |
Species & predicted species cross- reactivity ( ): | Human |
Applications & Suggested starting dilutions:* | WB 1:1000 IP n/d IHC 1:200 ICC n/d FACS n/d |
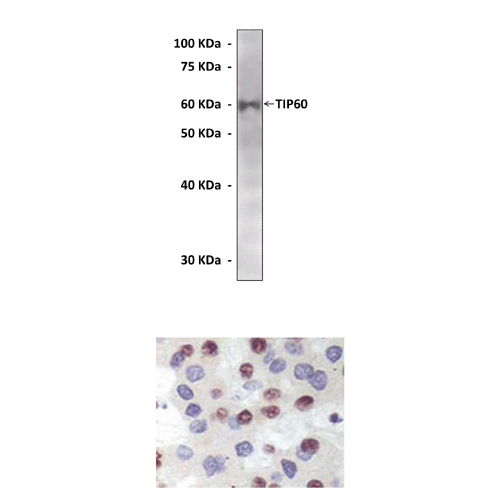
Predicted Molecular Weight of protein: | 60 kDa |
Specificity/Sensitivity: | Detects TIP60 proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Mouse Tat-Interactive Protein 60 Antibody: Mouse Tat-Interactive Protein 60 Antibody | Size: 100 ul | CAT.#: CP10237 | Price: $413.00 |