Product Sheet CP10432
Description
BACKGROUND The MCM (mini-chromosome maintenance proteins) proteins are essential replication initiation factors originally identified as proteins required for minichromosome maintenance in Saccharomyces cerevisiae. The best known among them are a family of six structurally related proteins, MCM2–7, which are evolutionally conserved in all eukaryotes. The MCM2–7 proteins form a hexameric complex with 1:1:1:1:1:1 stoichiometry and likely have a ring-shaped structure that surrounds DNA through its central channel. This complex is believed to function as the eukaryotic replicative DNA helicase. It plays important role in initiation of DNA replication.1 The initiation of DNA replication in eukaryotic cells is controlled by the stepwise establishment of prereplication complexes (pre-RCs) at DNA replication origins in G1 (also known as origin licensing) and the activation of two S phase–promoting kinases, Cdks/cyclins, and Cdc7/Dbf4, in G1/S. The pre-RCs contain several groups of proteins that are essential for the initiation of DNA replication. These include six subunits of origin recognition complex (ORC), the loading factors Cdc6 and Cdt1 proteins, and the putative DNA replicative helicase MCM2-7 complex. Origin licensing is sequential, with ORC binding to replication origins, which recruits the Cdc6 and Cdt1 proteins and thereby promotes the loading of MCM proteins. Although necessary, origin licensing is not sufficient to initiate DNA replication. The initiation of DNA replication requires the activation of two S phase–promoting kinases, Cdks/cyclins and Cdc7/Dbf4 in G1/S. It is thought that both S phase–promoting kinases phosphorylate critical downstream targets at pre-RCs that trigger the initiation of DNA replication. It was found that Cdc7/Dbf4 selectively phosphorylates MCM2 subunit (human MCM2 at three major sites: Ser27, Ser41, and Ser139 and two minor sites: Ser53 and Ser108) of MCM complex.2 In G1, when Cdc7/Dbf4 activity is at a minimum, MCM2 is not phosphorylated by Cdc7/Dbf4. Unphosphorylated MCM2 together with other MCMs (MCM2-7 complex) is recruited to chromatin, presumably to the replication origins by the DNA replication loading factors Cdc6 and Cdt1 to establish pre-RCs. During G1/S and early S, activation of Cdc7/Dbf4 results in phosphorylation of chromatin-bound MCM2. Phosphorylation of MCM2 in the MCM2-7 complex by Cdc7/Dbf4 induces the conformational change of the complex and activates its helicase activity, which is essential for DNA replication. MCM2-7 helicase works at a distance from the replicative forks, pumping DNA along its helical axis by ATPase-coupled rotation. In late S and G2/M, MCM2-7 complex dissociates from chromatin, presumably by additional post-translational modifications to prevent DNA rereplication.
After initiation of replication, MCM proteins dissociate from the origin, and this prevents a second round of DNA replication from the same origin during the same S phase. These proteins may also be required for unwinding the parental DNA strands during replication fork progression. Orchestration of the functional interactions between the MCM2–7 proteins and other components of the prereplication complex by cell cycle–dependent protein kinases results in initiation of DNA synthesis once every cell cycle,3 which is required for the entry in S phase and for cell division. New evidence suggests that the MCM2–7 proteins may be involved not only in the initiation but also in the elongation of DNA replication. Moreover, MCM2 is undetectable in quiescent cells and, thus, serves as a specific marker for replication-competent proliferating cells. It was suggested that that the use of MCM2 Antibodies is likely to provide a different, perhaps better, correlation with clinical outcome than the use of Anti-Ki-67 Antibodies.4
After initiation of replication, MCM proteins dissociate from the origin, and this prevents a second round of DNA replication from the same origin during the same S phase. These proteins may also be required for unwinding the parental DNA strands during replication fork progression. Orchestration of the functional interactions between the MCM2–7 proteins and other components of the prereplication complex by cell cycle–dependent protein kinases results in initiation of DNA synthesis once every cell cycle,3 which is required for the entry in S phase and for cell division. New evidence suggests that the MCM2–7 proteins may be involved not only in the initiation but also in the elongation of DNA replication. Moreover, MCM2 is undetectable in quiescent cells and, thus, serves as a specific marker for replication-competent proliferating cells. It was suggested that that the use of MCM2 Antibodies is likely to provide a different, perhaps better, correlation with clinical outcome than the use of Anti-Ki-67 Antibodies.4
REFERENCES
1. Kearsey, S.E. et al: BioEssays 18:183-190, 2005
2. Tsuji, T. et al:Mol. Biol. Cell 17:4459-72, 2006
3. Tye, B.K. et al: Ann Rev. Biochem. 68:649-86, 1999
4. Ramnath, N. et al : J. Clin. Oncol., 19: 4259-4266, 2001
2. Tsuji, T. et al:Mol. Biol. Cell 17:4459-72, 2006
3. Tye, B.K. et al: Ann Rev. Biochem. 68:649-86, 1999
4. Ramnath, N. et al : J. Clin. Oncol., 19: 4259-4266, 2001
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
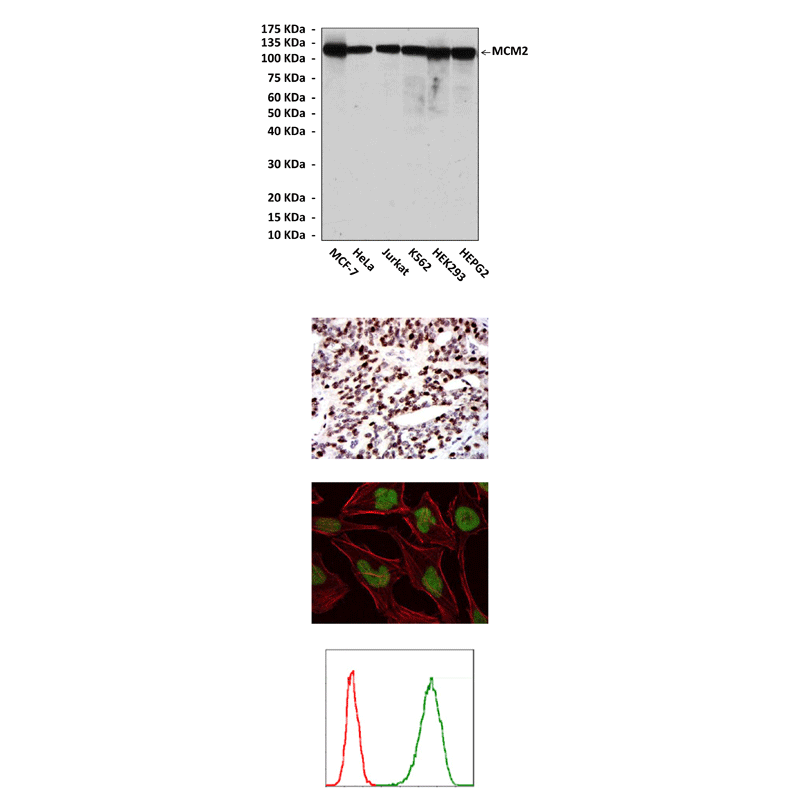
(Click to Enlarge) Top: Western blot detection of MCM2 proteins in various cell lysates using MCM2 Antibody. Middle, Upper: It also stains paraffin-embedded human ovarian cancer tissue in IHC analysis. Middle, Bottom: This antibody stains Hela cells in confocal immunofluorescent testing (MCM2 Antibody: Green; Actin filaments: Red). Bottom: This antibody also specifically detects MCM2 proteins in Jurkat cells in FACS assay (MCM2 Antibody: Green; control mouse IgG: Red).
Details
Cat.No.: | CP10432 |
Antigen: | Raised against recombinant human MCM2 fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
Applications & Suggested starting dilutions:* | WB 1:1000 IP 1:50 - 1:100 IHC 1:50 - 1:200 ICC 1:50 - 1:200 FACS 1:50 - 1:200 |
Predicted Molecular Weight of protein: | 125 kDa |
Specificity/Sensitivity: | Detects MCM2 proteins in various cell lysate. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Mouse MCM2 Antibody: Mouse MCM2 Antibody | Size: 100 ul | CAT.#: CP10432 | Price: $457.00 |
