Product Sheet CP10172
Description
BACKGROUND The mitotic checkpoint is a fail-safe mechanism that ensures accurate chromosome segregation by preventing cells from prematurely exiting mitosis in the presence of unaligned chromosomes. This checkpoint system is highly sensitive, because even a single unaligned chromosome is sufficient to block cells from entering anaphase. The mitotic checkpoint has been shown to monitor both microtubule attachment and tension generated across sister kinetochores by poleward forces. Failure of the mitotic checkpoint causes cells to exit mitosis in the presence of unaligned chromosomes and is a major mechanism responsible for aneuploidy. Seven mitotic checkpoint genes, BUB1, BUB2, BUB3, MAD1, MAD2, MAD3, and MPS1, were originally identified via genetic screens in Saccharomyces cerevisiae. These genes act along two separate mitotic checkpoint pathways. MPS1, BUB1, BUB3, MAD1, MAD2, and MAD3 monitor kinetochore microtubule attachments and prevent premature chromosome segregation by inhibiting degradation of securin/Pds1 and mitotic cyclins. BUB2 acts along a different pathway that monitors spindle integrity and orientation and prevents premature cytokinesis by inhibiting the degradation of the mitotic cyclin Clb2.
MPS1 was originally identified as a gene essential for spindle pole duplication and mutation in which yielded monopolar spindles. Vertebrate homologs of yeast MPS1 kinase have been identified as TTK (hMPS1) in humans and Esk (mMPS1) in mice. In all these MPS1 family members, the C-terminal catalytic domains show a significant degree of sequence similarity. However, the N-terminal domains show little, if any, sequence conservation. MPS1 is found to localize to kinetochores It encodes a tyrosine and serine/threonine dual-specificity kinase that was originally identified in a genetic screen for mutants defective in spindle pole duplication. Consistent with yeast MPS1, mouse MPS1 is localized at centrosomes throughout the cell cycle and is essential for accurate centrosome duplication. hMPS1 is required for the establishment and/or maintenance of the spindle assembly checkpoint in human cells.1 hMPS1 was also found to be required for centrosome duplication and for the normal progression of mitosis.2 Furthermore, xMPS1 was found to be critical for the localization of CENP-E and the checkpoint proteins MAD1 and MAD2 to kinetochores. Likewise, disruption of hMPS1 prevented cells from arresting in mitosis in the presence of spindle damage. In addition, hMPS1 is hyperphosphorylated in mitosis and is dephosphorylated when cells exit mitosis. hMPS1, as was shown for human MAD1 and MAD2, also localizes to the nucleoplasmic side of the nuclear pore complex (NPC). Furthermore, hMPS1 is part of the checkpoint pathway that is required to arrest cells defective for CENP-E functions in mitosis. In addition to its localization at kinetochores, it was found hMPS1 to associate with the APC. This suggests that hMPS1 may have multiple roles in the mitotic checkpoint.3 Mps1 phosphorylation by MAPK at S844 might create a phosphoepitope that allows Mps1 to interact with kinetochores. Recently, it was demonstrated Mps1 kinase activity is essential for chromosome alignment by enhancing Aurora B activity at the centromere, and the Aurora B-regulatory protein Borealin/DasraB is an essential substrate that mediates this novel function of MPS1.4 In addition, Mps1 Phosphorylation of Dam1 Couples Kinetochores to Microtubule plus Ends at Metaphase.5
MPS1 was originally identified as a gene essential for spindle pole duplication and mutation in which yielded monopolar spindles. Vertebrate homologs of yeast MPS1 kinase have been identified as TTK (hMPS1) in humans and Esk (mMPS1) in mice. In all these MPS1 family members, the C-terminal catalytic domains show a significant degree of sequence similarity. However, the N-terminal domains show little, if any, sequence conservation. MPS1 is found to localize to kinetochores It encodes a tyrosine and serine/threonine dual-specificity kinase that was originally identified in a genetic screen for mutants defective in spindle pole duplication. Consistent with yeast MPS1, mouse MPS1 is localized at centrosomes throughout the cell cycle and is essential for accurate centrosome duplication. hMPS1 is required for the establishment and/or maintenance of the spindle assembly checkpoint in human cells.1 hMPS1 was also found to be required for centrosome duplication and for the normal progression of mitosis.2 Furthermore, xMPS1 was found to be critical for the localization of CENP-E and the checkpoint proteins MAD1 and MAD2 to kinetochores. Likewise, disruption of hMPS1 prevented cells from arresting in mitosis in the presence of spindle damage. In addition, hMPS1 is hyperphosphorylated in mitosis and is dephosphorylated when cells exit mitosis. hMPS1, as was shown for human MAD1 and MAD2, also localizes to the nucleoplasmic side of the nuclear pore complex (NPC). Furthermore, hMPS1 is part of the checkpoint pathway that is required to arrest cells defective for CENP-E functions in mitosis. In addition to its localization at kinetochores, it was found hMPS1 to associate with the APC. This suggests that hMPS1 may have multiple roles in the mitotic checkpoint.3 Mps1 phosphorylation by MAPK at S844 might create a phosphoepitope that allows Mps1 to interact with kinetochores. Recently, it was demonstrated Mps1 kinase activity is essential for chromosome alignment by enhancing Aurora B activity at the centromere, and the Aurora B-regulatory protein Borealin/DasraB is an essential substrate that mediates this novel function of MPS1.4 In addition, Mps1 Phosphorylation of Dam1 Couples Kinetochores to Microtubule plus Ends at Metaphase.5
REFERENCES
1. Stucke,V.M. et al: EMBO J. 21:1723-32, 2002
2. Fisk, H.A. et al: Proc. Natl Acad. Sci. USA 100:14875-80, 2003
3. Liu, S-T. et al: Mol. Biol. Cell 14:1638-51, 2003
4. Jelluma, N. et al: Cell 132:23-46, 2009
5. Shimogawa, M.M. et al: Curr. Biol. 16:1489-1501, 2006
2. Fisk, H.A. et al: Proc. Natl Acad. Sci. USA 100:14875-80, 2003
3. Liu, S-T. et al: Mol. Biol. Cell 14:1638-51, 2003
4. Jelluma, N. et al: Cell 132:23-46, 2009
5. Shimogawa, M.M. et al: Curr. Biol. 16:1489-1501, 2006
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Details
Cat.No.: | CP10172 |
Antigen: | Purified recombinant human MPS1 fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
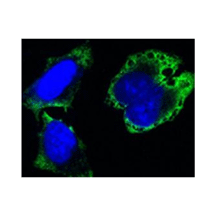
Applications & Suggested starting dilutions:* | WB n/d IP n/d IHC n/d ICC 1:200 FACS n/d |
Predicted Molecular Weight of protein: | 95 kDa |
Specificity/Sensitivity: | Detects endogenous MPS1 proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Mouse Monopolar Spindle-1 Antibody: Mouse Monopolar Spindle-1 Antibody | Size: 100 ul | CAT.#: CP10172 | Price: $413.00 |