Product Sheet CP10283
Description
BACKGROUND Circadian rhythms of both the central nervous system and peripheral tissues and organs are generated by the coordinated activation/inactivation of self-oscillating transcription factors. Central among them are CLOCK and its heterodimer partner brain-muscle-arnt-like protein 1 (BMAL1), which belong to the basic helix-loop-helix (bHLH)-PER-ARNT-SIM (PAS) superfamily of transcription factors. The CLOCK/BMAL1 heterodimer stimulates the transcription of other essential clock genes, such as the Period (PER1, PER2, and PER3) and Cryptochrome (CRY1 and CRY2) genes. PER and CRY proteins, then, suppress their own transcription by repressing the transcriptional activity of CLOCK/BMAL1, in essence forming a self-oscillating, negatively regulated feedback loop system. MOP4 [also called neuronal PAS domain protein 2 (NPAS2)] shares a high amino acid homology with CLOCK, forms heterodimers with BMAL1, and also participates in controlling the regulatory loop of circadian oscillator machinery. Moreover, it was shown that CLOCK phosphorylation contributes to the suppression of CLOCK/BMAL1-mediated transactivation through dual regulation: inhibition of CLOCK activity and promotion of its degradation. Furthermore, it was shown that receptor for activated C kinase–1 (RACK1) and protein kinase C–α (PKCα) were recruited in a circadian manner into a nuclear BMAL1 complex during the negative feedback phase of the cycle. Thus, the classical PKC signaling pathway is rhythmically activated by internal processes, forming an integral part of the circadian feedback loop.1 Interestingly, CLOCK also shares high amino acid and structural similarity with the activator of thyroid receptor (ACTR), a member of the p160-type nuclear receptor coactivator family with inherent HAT activity, and thus, like these proteins, has such enzymatic function, without which CLOCK/BMAL1 is unable to generate a circadian rhythm. CLOCK and MOP4 interact with the nuclear receptor family members retinoic acid receptor (RAR)-α and retinoic X receptor (RXR)-α, which negatively regulate CLOCK/BMAL1-mediated transcriptional activity of clock gene expression. In addition, it was demonstrated that CLOCK/BMAL1 regulates glucocorticoid actions in peripheral tissues by directly interacting with and enzymatically targeting the glucocorticoid receptor (GR). CLOCK/BMAL1 acetylates the GR at a cluster of lysine residues in its hinge region and represses GR-induced transcriptional activity by attenuating the association of GR to GREs.2
The complex program of gene expression that characterizes circadian physiology is possible through dynamic changes in chromatin transitions. These remodeling events are therefore of great importance to insure the proper timing and extent of circadian regulation. Recent advances in the field have revealed unexpected links between circadian regulators, chromatin remodeling and cellular metabolism. Specifically, the central clock protein CLOCK has HAT enzymatic properties. It directs acetylation of histone H3 and of its dimerization partner BMAL1 at K537, an event essential for circadian function. In addition, the HDAC activity of the NAD+-dependent SIRT1 enzyme is regulated in a circadian manner. It has been proposed that SIRT1 functions as an enzymatic rheostat of circadian function, transducing signals originated by cellular metabolites to the circadian clock. Thus, a specialized program of chromatin remodeling appears to be at the core of the circadian machinery.3 Emerging evidence suggests a close link between the circadian clock system and metabolic homeostasis. It was shown that KLF10 (also known as Tieg1), previously shown to be a regulator of bone physiology, is a CLOCK/BMAL1-controlled transcription factor regulating genes implicated in glucose and lipid metabolism in liver.4 Finally, CLOCK is shown to be mutated in cancer, and altered response to DNA damage provides one plausible mechanism of tumorigenesis.5
The complex program of gene expression that characterizes circadian physiology is possible through dynamic changes in chromatin transitions. These remodeling events are therefore of great importance to insure the proper timing and extent of circadian regulation. Recent advances in the field have revealed unexpected links between circadian regulators, chromatin remodeling and cellular metabolism. Specifically, the central clock protein CLOCK has HAT enzymatic properties. It directs acetylation of histone H3 and of its dimerization partner BMAL1 at K537, an event essential for circadian function. In addition, the HDAC activity of the NAD+-dependent SIRT1 enzyme is regulated in a circadian manner. It has been proposed that SIRT1 functions as an enzymatic rheostat of circadian function, transducing signals originated by cellular metabolites to the circadian clock. Thus, a specialized program of chromatin remodeling appears to be at the core of the circadian machinery.3 Emerging evidence suggests a close link between the circadian clock system and metabolic homeostasis. It was shown that KLF10 (also known as Tieg1), previously shown to be a regulator of bone physiology, is a CLOCK/BMAL1-controlled transcription factor regulating genes implicated in glucose and lipid metabolism in liver.4 Finally, CLOCK is shown to be mutated in cancer, and altered response to DNA damage provides one plausible mechanism of tumorigenesis.5
REFERENCES
1. Robles, M.S. et al: Science 327:463-6, 2010
2. Nader, N. et al: FASEB J. 23:1572-83, 2009
3. Grimaldi, B. et al: Int. J. Biochem. Cell Biol. 41:81-6, 2009
4. Guillaumond, F. et al: Mol. Cell. Biol. 30:3059-70, 2010
5. Alhopuro, P. et al: Mol. Cancer Res. 8: 952–60, 2010
2. Nader, N. et al: FASEB J. 23:1572-83, 2009
3. Grimaldi, B. et al: Int. J. Biochem. Cell Biol. 41:81-6, 2009
4. Guillaumond, F. et al: Mol. Cell. Biol. 30:3059-70, 2010
5. Alhopuro, P. et al: Mol. Cancer Res. 8: 952–60, 2010
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
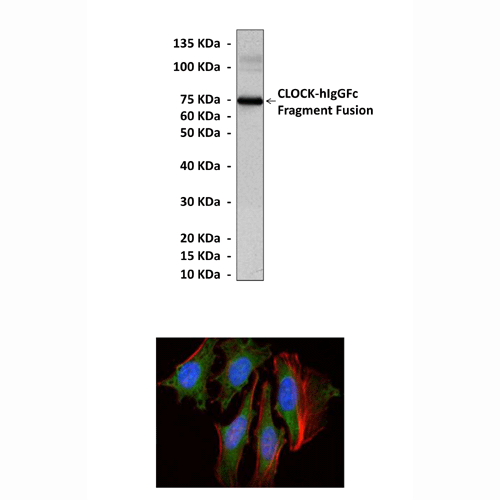
(Click to Enlarge) Top: Western Blot detection of CLOCK proteins in 293 cells expressing recombinant human CLOCK fragment-hIgGFc fusion proteins using CLOCK Antibody. Bottom: This antibody stains HeLa cells in confocal immunofluorescent analysis (CLOCK Antibody: Green; Actin filaments: Red; DRAQ5 DNA Dye: Blue).
Details
Cat.No.: | CP10283 |
Antigen: | Raised against recombinant human CLOCK fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human |
Applications & Suggested starting dilutions:* | WB 1:1000 IP n/d IHC n/d ICC n/d FACS n/d |
Predicted Molecular Weight of protein: | 95 kDa |
Specificity/Sensitivity: | Detects CLOCK proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Products
| Product | Size | CAT.# | Price | Quantity |
|---|---|---|---|---|
| Mouse CLOCK Antibody: Mouse CLOCK Antibody | Size: 100 ul | CAT.#: CP10283 | Price: $354.00 |
